Overview
Introduction
Computation plays an important role in materials science, engineering, nanotechnology, pharmaceutical research and many other research fields. Faster time to market, increased return on investment, and enabling new products are common reasons computation is used in product development. However, surveys have shown that moderate sized companies are only slowly adopting materials simulations and taking advantage of the potential increases in innovation and productivity.
Improving the adoption of computational chemistry software is addressed by developing a framework to help streamline complex simulation workflows involving one or more of the following: molecular dynamics, quantum chemistry simulations, molecular docking, kinetic Monte-Carlo and others. Chemistream is a user-friendly application capable of taking advantage of cloud computing resources, using the sub-package Makalii and combining HPC simulations into industry workflows using the STREAMM package developed at NREL.
Chemistream is a framework for managing complex HPC materials simulation workflows built on a JupyterLab framework. The philosophy of Chemistream is to use open-source Python packages and open-source simulation engines developed at national labs and the DOE to provide HPC resources in the cloud for small- and medium-sized industries. By making cutting-edge, open-source software from the national labs easier to use, their entire user communities, including those in academia and at the national labs, will benefit as well.
- We are seeking to improve simulations for
innovation in organic electronics (batteries, solar cells)
the product development cycle in materials processing companies
materials involved in photonics
training AI/ML models for materials discovery
Design
Chemistream combines the Makalii python package (developed by Tech-X), the STREAMM python package (developed by NREL), and other workflow/visualization/etc modules with HPC cloud containers (developed by Tech-X and UberCloud) in a user-friendly JupyterLab framework. The hierarchical design of Chemistream allows users to take advantage of its functionality at a level that is most comfortable for them:
Command-line Users Chemistream can be used as simply a way to manage a remote cluster and install the desired software. Users can then login to their remote cluster on the command-line in much the same way as logging on to an HPC cluster at NERSC or a national lab
Workflow Users Chemistream can launch remote notebooks in the cloud that implement a workflow that is tailored to their simulation/modeling needs. These workflows manage cloud resources, as well as simulation job setup, monitoring and analysis
Power Users Chemistream is essentially a toolkit. Power users use any part of this toolkit in a remote HPC cloud computing environment and take advantage of the library level functionality provided by STREAMM and proprietary Chemistream code. In short, ‘Power Users’ can write their own workflows.
All of the above The Chemistream interface allows a user to have a command line terminal, pre-written workflows and user-defined workflows running all at the same time. Mix and match the levels to meet your needs.
HPC Cloud Images from Tech-X
The Chemistream application can setup remote clusters with Docker images pre-configured with simulation codes and analysis/visualization environments. All of the images contain the environment that enables the remote Chemistream framework. The current selection of images includes:
Alfabet: Image including the base-level Chemistream environment and the ALFABET (link) module that uses artificial intelligence (AI) to predict bond-dissociation energies. Also includes the JSME molecular drawer program and the Tensorflow package.
GROMACS: Image including the base-level Chemistream environment and the classical molecular dynamics code GROMACS. This image enables networked remote compute clusters and configures a SLURM scheduler.
LAMMPS-multi: Image including the base-level Chemistream environment and classical molecular dynamics code LAMMPS. This image enables networked remote compute clusters and configures a SLURM scheduler.
NWChem-multi: Image including the base-level Chemistream environment and DFT quantum chemistry code NWChem. This image enables networked remote compute clusters and configures a SLURM scheduler.
NWChem+DDEC6-multi: Image including the base-level Chemistream environment, the DFT quantum chemistry code NWChem and the charge analysis code DDEC6. This image enables networked remote compute clusters and configures a SLURM scheduler.
NWChem+LAMMPS-multi: Image including the base-level Chemistream environment, the DFT quantum chemistry code NWChem and the classical molecular dynamics code LAMMPS. This image enables networked remote compute clusters and configures a SLURM scheduler.
QMCPack-multi: Image including the base-level Chemistream environment and the quantum Monte-Carlo code QMCPack. This image enables networked remote compute clusters and configures a SLURM scheduler.
Dalton-multi: Image including the base-level Chemistream environment and the DFT quantum chemistry code Dalton. This image enables networked remote compute clusters and configures a SLURM scheduler.
rDock: Image including the base-level Chemistream environment and the rDock molecular docking program.
RLMolecule: Image including the base-level Chemistream environment and the RLMolecule module that uses reinforcement learning (RL) to direct molecular design. Also includes the JSME molecular drawer program and the Tensorflow package.
SPPARKS-multi: Image including the base-level Chemistream environment and the kinetic Monte-Carlo code SPPARKS. This image enables networked remote compute clusters and configures a SLURM scheduler.
SPPARKS-multi-ALD: Image including the base-level Chemistream environment and the kinetic Monte-Carlo code SPPARKS compiled with a model for Atomic Layer Deposition (ALD). This image enables networked remote compute clusters and configures a SLURM scheduler.
Tensorflow-(CPU)GPU: Image including the base-level Chemistream environment and the Tensorflow package with a custom configuration needed to enable GPUs for neural network training.
Remote Cluster Setup
Chemistream uses a custom, proprietary image base layer (developed w/Ubercloud) to create HPC, networked remote clusters on cloud resources. The setup is general and portable across cloud providers and is currently available on Amazon Web Services, Azure and for private clouds. The Chemistream interface manages all of the complexities of service discovery across separate compute instances, setting up a shared NFS directory and configuring Slurm scheduling, thereby providing a ‘cluster-on-the-fly’.
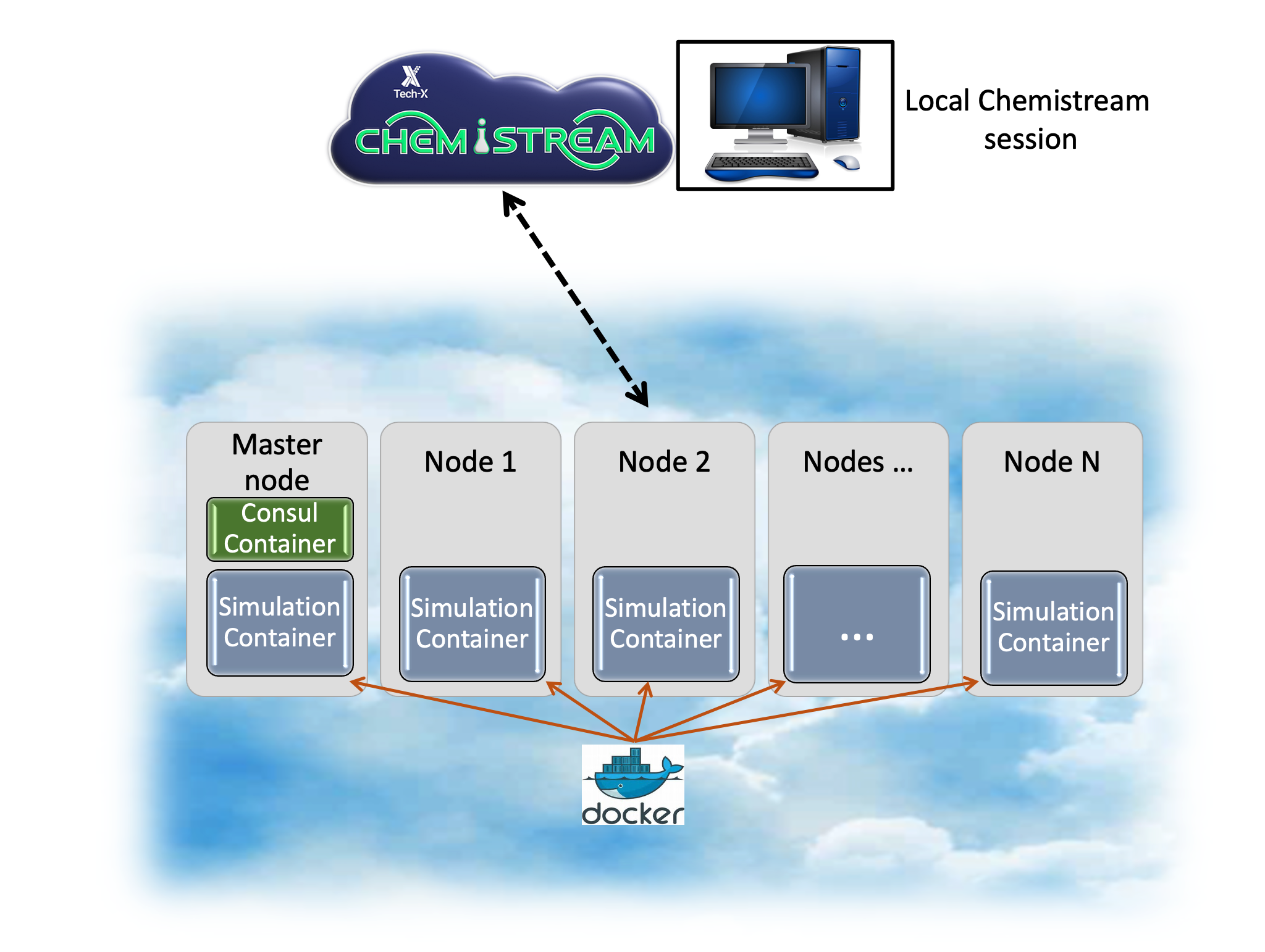
The following steps are managed through the Chemistream application and are shown in detail in the tutorials.
create remote instances (e.g. AWS, private cluster, Azure[coming soon])
remote copy setup scripts and configuration files (e.g. for Slurm, NFS shared directory etc.)
remote install setup programs, Docker etc.
remote pull Consul service-discovery image on the master node
remote pull user-specified simulation Docker images (see HPC Cloud Images from Tech-X)
remote start containers
Source Code Licensing Agreements
These source code licensing agreements are also available in compute sessions. Source code is accessible in compute sessions for codes that have GPL2 licensing scheme.